1.4 - Installing and activating Variantseq and the Server-Side
VariantSeq is a Client-Side + Server Side application. You can download the latest version of VariantSeq (the client side) at the following URL https://gpro.biotechvana.com/download/VariantSeq.
To install VariantSeq on your PC, follow
the instructions in the manual at https://gpro.biotechvana.com/tool/variantseq/manual. To install this app, you must have Java 11 previously
installed on your PC.
The GPRO Server Side can be installed on a PC or a remote server as a Cloud Computing resource. However, due to the complexity of installing the program, we distribute the GPRO Server Side in a Docker container (Merkel 2014) The Docker container can be installed easily by following the steps described here: https://gpro.biotechvana.com/tool/gpro-server/manual.
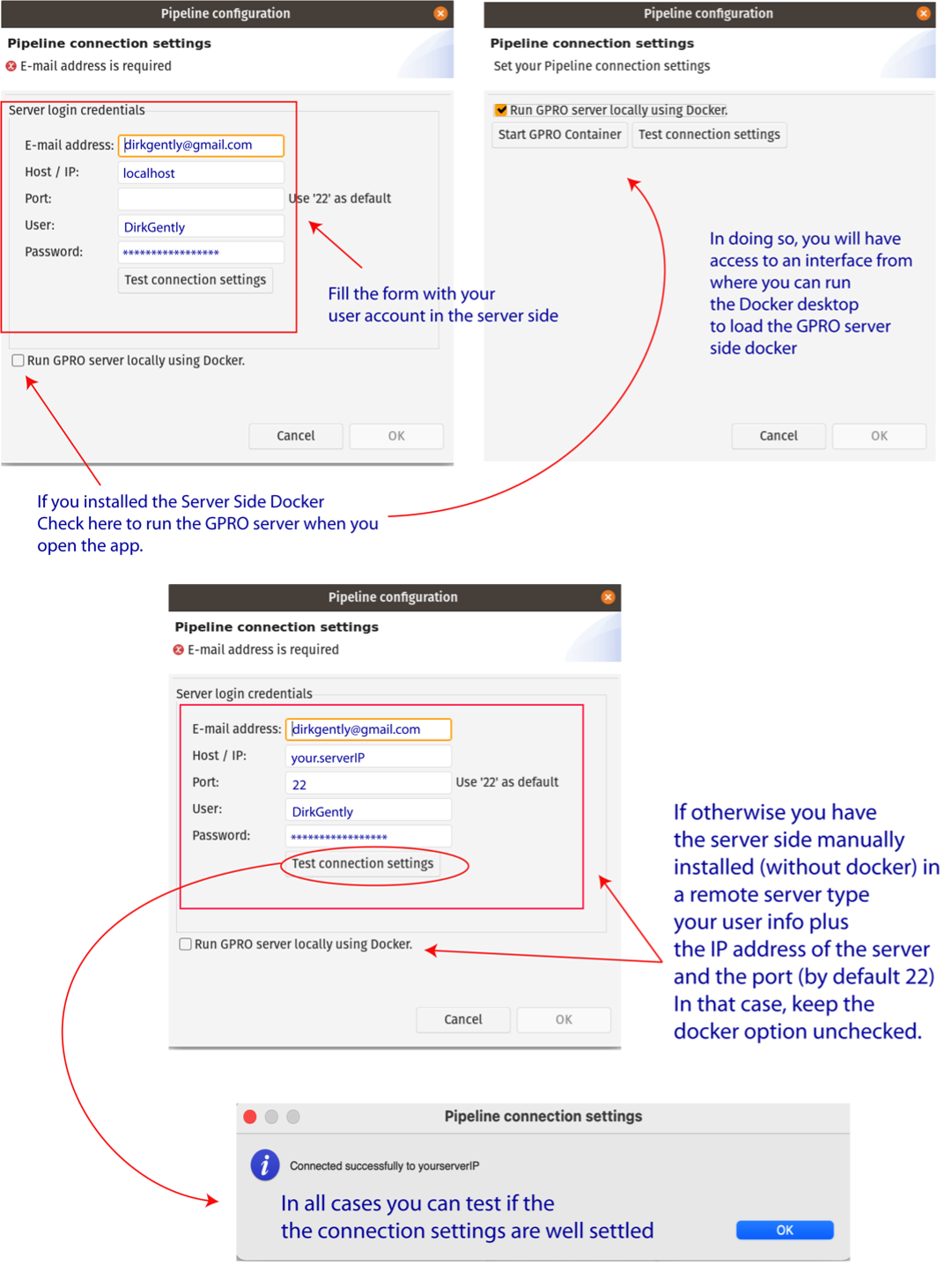
Once the GPRO server side docker has been installed, it must be linked to VariantSeq. To do this, go to [Preferences → Pipeline connection settings] in the top menu and type the following into the configuration Dialog (Fig.1):
- Your email address: to receive notifications from the server.
- Host / IP address: here you should type localhost (see Fig.1).
- Port: This field should be filled only in case you run the server-side manually. The default number will be 22.
- Username and password: Your ID credentials provided to access the host server.
As shown in Fig.1, you can also check the option “Run GPRO server locally using Docker” to automatically start the GPRO container each time you run VariantSeq (if you have this option checked, you do not need to type in the port number). You can test if the app is connected to the Server Side by clicking on the tab “Test connection settings”. Alternatively, if you have installed the Server Side manually (without the Docker), add the IP of the remote server where the Server Side is hosted and the port information (by default 22). You should also leave the Option “Run GPRO server locally using Docker” unchecked.